What are the developments?
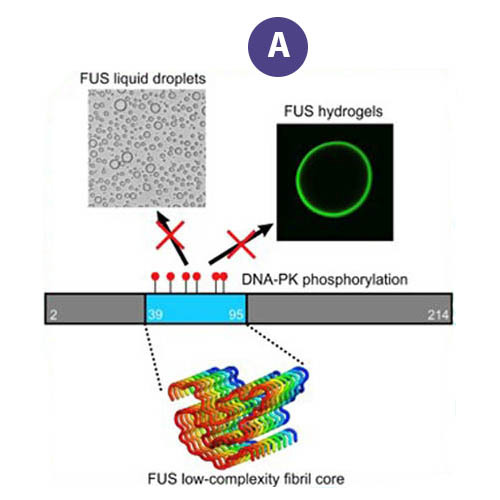
Data users from Harvard and the University of Zurich accessed FAIR data from the Protein Data Bank generated by the MagLab's NMR Facility, data that describes the molecular structure of the RNA-binding protein, "FUS" (Figure A). The data were used to examine the effects of naturally-occurring mutations that change the structure of its fibril core among mammals dating back 160 million years.
Why is this important?
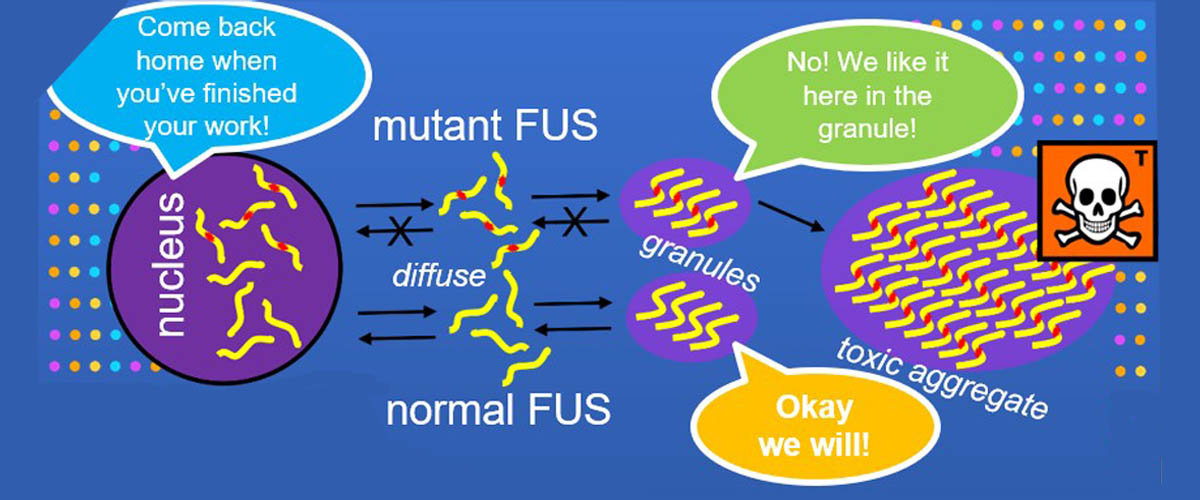
Aggregation of the protein "FUS" is associated with several neurodegenerative diseases. These proteins form liquid compartments, or "granules", where they perform a variety of vital functions. NMR experiments revealed that formation of these granules is controlled by the way the FUS fibril core domain is folded. Normally, the granules are short-lived, but mutations in FUS cause the core domain to permanently adopt the granule-favored structure. Over time, they accumulate and form toxic aggregates (Figure B) — a hallmark of ALS. Evolutionary biologists accessed the NMR data to explore ways that evolution and natural selection have influenced the structure of FUS and its aggregate forming potential (Figure C). This helps to identify and understand evolutionary patterns of disease-causing proteins and suggests new strategies for deepening our understanding of important biological systems so that we can improve our quality of life.
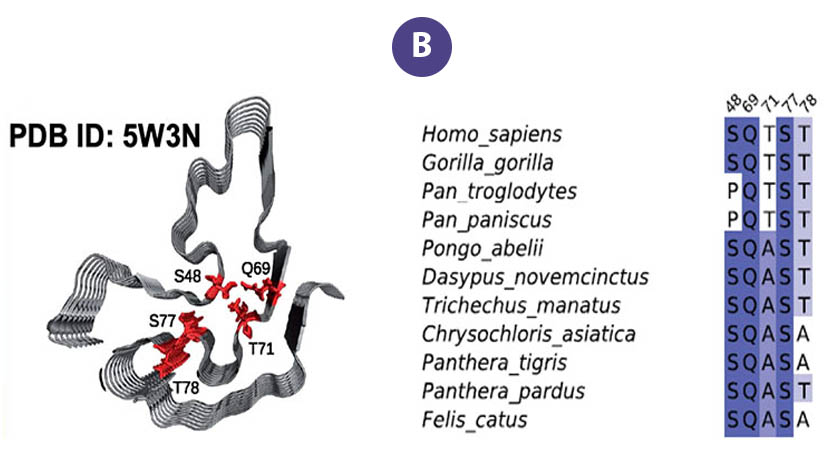
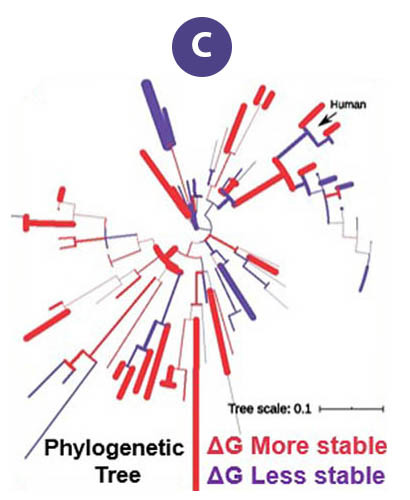
Image credit: Lissa C. Anderson
Why did they need the MagLab?
The original data taken on the MagLab’s world-unique 900MHz ultrawide bore magnet served as a scaffold upon which evolutionary biologists could model and study the RNA-binding protein "FUS" from other species, living and extinct.
Details for scientists
- View or download the expert-level Science Highlight, NMR FAIR Data - Effects of Natural Selection on the Phase-Separation Properties of an RNA-Binding Protein in Mammals
Funding
Original data collected under the MagLab’s NSF/DMR-1644779
Data User research was funded by grants awarded to Pouria Dasmeh1,2,3, Andreas Wagner1,3, European Research Council #739874, Swiss NSF #31003A 172887.
1Institute for Evolutionary Biology and Environmental Studies, University of Zurich; 2Dept. of Chemistry and Chemical Biology, Harvard University; 3Swiss Institute of Bioinformatics
For more information, contact Lissa C. Anderson.