What did scientists discover?
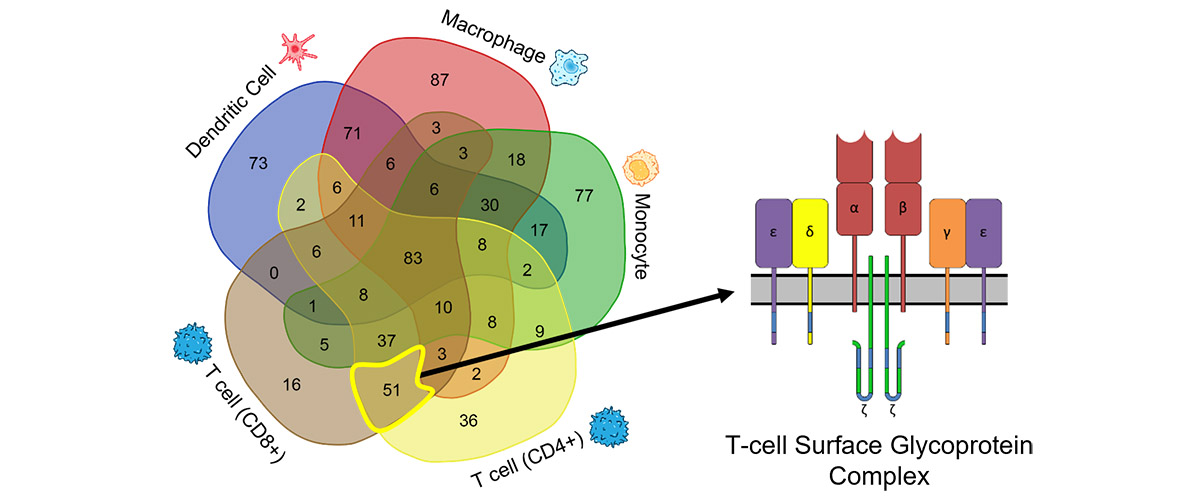
Melani et al. compiled a Blood Proteoform Atlas (BPA), a clarified map of ~30,000 distinct molecular forms of proteins, or "proteoforms", as they appear in 21 different blood cell types. With the BPA as a reference, this team of MagLab users, headed by Melani, examined proteoforms in patient blood samples and developed a panel of 24 biomarkers for liver transplant rejection.
Why is this important?
Through employment of state-of-the-art instrumentation, including the MagLab's 21T FT-ICR mass spectrometer, proteoforms were identified intact in a form of "top-down" analysis rather than cutting them into small pieces (as is standard practice). This advancement realizes a truly molecular-level understanding of cell type and marks the beginning of a new era for more precise study of proteins in specific cells— the Human Proteoform Project. Using the Atlas as a reference, researchers examined proteoforms in patient blood samples and developed a panel of biomarkers for liver transplant rejection.
Who did the research?
Rafael D. Melani1, Vincent R. Gerbasi1, Lissa C. Anderson2, et al.
1Northwestern University; 2National High Magnetic Field Laboratory
Why did they need the MagLab?
High-throughput identification of proteoforms from complex biological samples requires instrumentation capable of high mass resolving power, mass accuracy, sensitivity, and efficiency. The 21 tesla FT-ICR mass spectrometer is state-of-the-art in all these facets and contributed nearly a third of the BPA's proteoforms while accounting for only ~15% of the total instrument time devoted to the project.
Details for scientists
- View or download the expert-level Science Highlight, The Blood Proteoform Atlas: A reference map of proteoforms in human blood cells
- Read the full-length publication The Blood Proteoform Atlas: A reference map of proteoforms in human hematopoietic cells, in Science
Funding
This research was funded by the following grants: G.S. Boebinger (NSF DMR-1644779); Neil L. Kelleher (Paul G. Allen Frontiers Program award 11715; HuBMAP grant UH3 CA246635-02; NIH P41 GM108569); and others
For more information, contact Christopher Hendrickson.