What is the finding
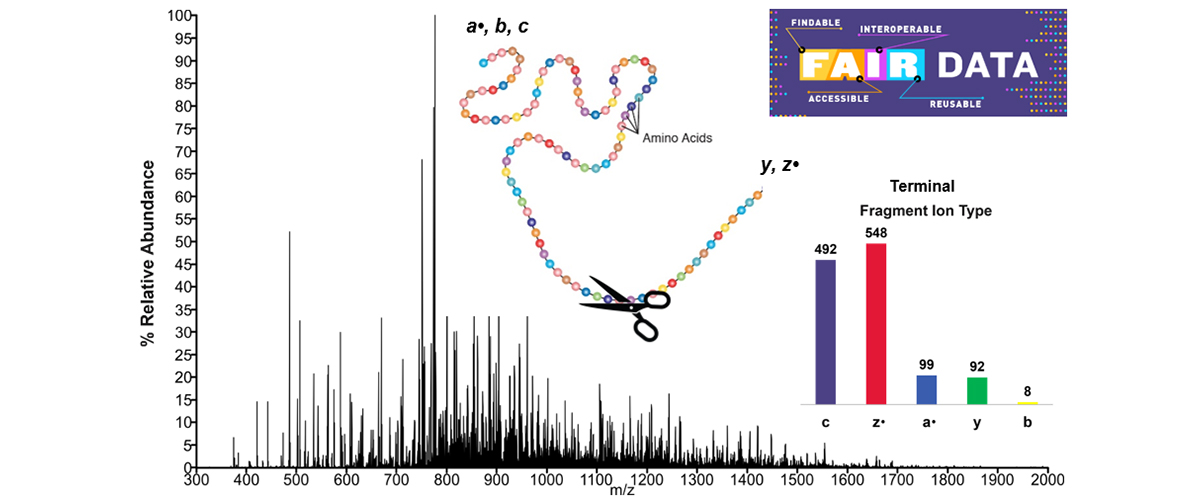
In mass spectrometry of intact proteins, electron-based fragmentation does not reveal internal fragment ions (Figure 1). Researchers used data from the MagLab’s 21 tesla FT-ICR mass spectrometer and other lower-resolution data from other labs. Automated software incorrectly identified internal fragments, but careful manual analysis proved these assignments were wrong.
Why is this important?
Electron-based fragmentation in mass spectrometry provides better protein sequence information than traditional methods by creating more observable terminal fragments. Adding internal fragments, formed by breaking multiple bonds, could improve coverage of the protein's interior, making it easier to identify important modifications that affect protein function in health and disease. However, misidentifying these fragments can lead to errors in the analysis.
Who did the research?
Neven N. Mikawy1, Carolina Rojas Ramírez1,2, Alexey I. Nesvizhskii,2 Joseph A. Loo3, Brandon T. Ruotolo1, Jeffrey Shabanowitz4, Lissa C. Anderson5, Kristina Håkansson1
Departments of 1Chemistry and 2Pathology, U. Michigan; 3Department of Chemistry and Biochemistry, UCLA; 4Department of Chemistry, U. Virginia; 5National MagLab (and others)
Why did they need the MagLab?
The 21 T FT-ICR mass spectrometer offers exceptional mass resolving power, mass accuracy, and sensitivity. These performance characteristics significantly enhance the confidence in assigning both terminal and internal fragments in complex mass spectra (Figure 1). In particular, low-abundance and high-mass fragments, which may not be observed or may be impossible to annotate in lower resolution data, can be confidently assigned, highlighting the risk associated with internal fragment annotation in mass spectra.
Details for scientists
- View or download the expert-level Science Highlight, Are Internal Fragments Observable in Electron Based Top-Down Mass Spectrometry?
- Read the full-length publication, Are Internal Fragments Observable in Electron Based Top-Down Mass Spectrometry?, in J. Am. Soc. Mass Spectrom Data Set
Funding
This research was funded by the following grants: K. M. Amm (NSF DMR-1644779 and DMR-2128556); K. Håkansson (NSF CHE-2004043, NIH S10OD021619); A. I. Nesvizhskii (NIH T32CA140044, R01GM094231, and U24CA271037); J. A. Loo (NIH R35GM145286 and DOE DE-FC02-02ER63421); B. T. Ruotolo (Agilent Technologies); D. F. Hunt (NIH GM037537))
For more information, contact Lissa Anderson.